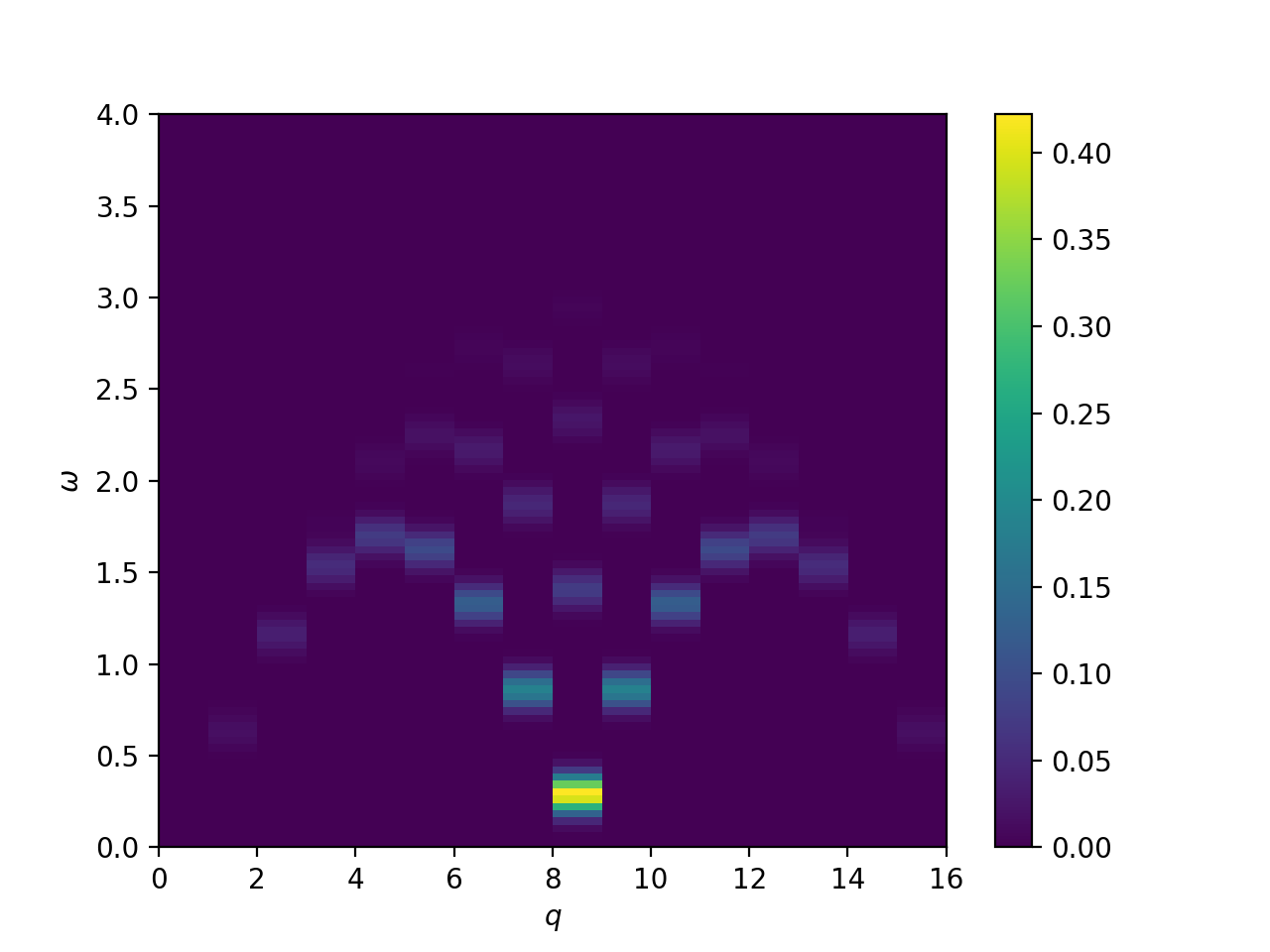
Dynamical structure factor of the Heisenberg S=1/2 chain
We compute the ground state energy of the \(S=1/2\) Heisenberg chain with periodic boundary conditions in each momentum sector \(k\). The Hamiltonian is given by
\[H = J\sum_{\langle i,j \rangle} \mathbf{S}_i \cdot \mathbf{S}_j\]where \(\mathbf{S}_i = (S_i^x, S_i^y, S_i^z)\) are the spin \(S=1/2\) operators and \(\langle i,j \rangle\) denotes summation over nearest-meighbor sites \(i\) and \(j\). We set \(J=1\).
First, the Lanczos coefficients are computed using hydra and written to file
#include <hydra/all.h>
int main() {
using namespace hydra;
using namespace arma;
int n_sites = 16;
int n_up = n_sites / 2;
// Create nearest-neighbor Heisenberg model
BondList bonds;
for (int s = 0; s < n_sites; ++s) {
bonds << Bond("HB", "J", {s, (s + 1) % n_sites});
}
bonds["J"] = 1.0;
// compute groundstate (known to be at k=0)
Log("Computing ground state ...");
auto block = Spinhalf(n_sites, n_up);
auto gs = groundstate(bonds, block);
Log("done.");
for (int q = 0; q < n_sites; ++q) {
Log("Dynamical Lanczos iterations for q={}", q);
complex phase = exp(2i * pi * q / n_sites);
BondList S_of_q;
for (int s = 0; s < n_sites; ++s) {
S_of_q << Bond("SZ", pow(phase, s) / n_sites, s);
}
// Compute S(q) |g.s.>
auto v0 = zero_state(block);
apply(S_of_q, gs, v0);
double nrm = norm(v0);
v0 /= nrm;
// Perform 200 Lanczos iterations for dynamics starting from v0
auto tmat = lanczos_eigenvalues_inplace(bonds, block, v0.vector(), 0, 0.,
200, 1e-7);
auto alphas = tmat.alphas();
auto betas = tmat.betas();
// Write alphas, betas, and norm to file for further processing
alphas.save(
fmt::format("outfiles/alphas.N.{}.nup.{}.q.{}.txt", n_sites, n_up, q),
raw_ascii);
betas.save(
fmt::format("outfiles/alphas.N.{}.nup.{}.q.{}.txt", n_sites, n_up, q),
raw_ascii);
std::ofstream of;
of.open(
fmt::format("outfiles/norm.N.{}.nup.{}.q.{}.txt", n_sites, n_up, q));
of << nrm;
of.close();
}
return EXIT_SUCCESS;
}
The produced data is the read from file and can be visualized using the following python script.
#!/usr/bin/env python
import numpy as np
import matplotlib.pyplot as plt
n_sites = 16
n_up = n_sites // 2
eta = 0.05
n_omegas = 100
max_omega = 4.0
omegas = np.linspace(0, max_omega, n_omegas)
# compute broadened spectrum
spectra = np.zeros((n_sites, n_omegas))
for q in range(n_sites):
alphas = np.loadtxt("outfiles/alphas.N.{}.nup.{}.q.{}.txt".format(n_sites, n_up, q))
betas = np.loadtxt("outfiles/betas.N.{}.nup.{}.q.{}.txt".format(n_sites, n_up, q))
norm = np.loadtxt("outfiles/norm.N.{}.nup.{}.q.{}.txt".format(n_sites, n_up, q))
# compute poles and weights from tridiagonal matrix
tmat = np.diag(alphas[:50]) + np.diag(betas[:49], k=1) + np.diag(betas[:49], k=-1)
eigs, evecs = np.linalg.eigh(tmat)
e0 = eigs[0]
poles = eigs - e0
weights = (norm**2) * (evecs[0, :]**2)
print(norm, e0)
# compute broadened spectrum with Gaussian broadening eta
diffs = np.subtract.outer(omegas, poles)
gaussians = np.exp(-(diffs / (2*eta))**2) / (eta * np.sqrt(2*np.pi))
spectra[q, :] = gaussians @ weights
p = plt.imshow(spectra.T, origin='lower', extent=[0, n_sites, 0, max_omega],
interpolation=None, aspect='auto')
plt.xlabel(r"$q$")
plt.ylabel(r"$\omega$")
plt.colorbar(p)
plt.show()
This will plot the following image: